When I wrote the draft of the manuscript entitled “Virtual screening of peptides with high affinity for SARS-CoV-2 main protease”, the pandemic of severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) had caused in about of 900,000 deaths worldwide. Today, this number is about to reach 3,000,000 of deaths, and Brazil has been considered as the new epicenter of the disease.
Fortunately, the vaccine development was accelerated and by now there are some options available. In fact, the vaccines are the ultimate resource to solve the pandemic. However, they are not the cure, but the prevention. Therefore, the search of new drugs to treat the coronavirus disease 2019 (COVID-19) are still valid, in particular for those people which are hospitalized.
In this context, peptides have been poorly explored as a potential drug for COVID-19. The main strategy has been the repositioning of drugs already approved for human use, where the virtual screening plays a pivotal role, exploring hundreds to thousands of molecules. In this context, I developed a virtual screening system for peptides against the viral protease.
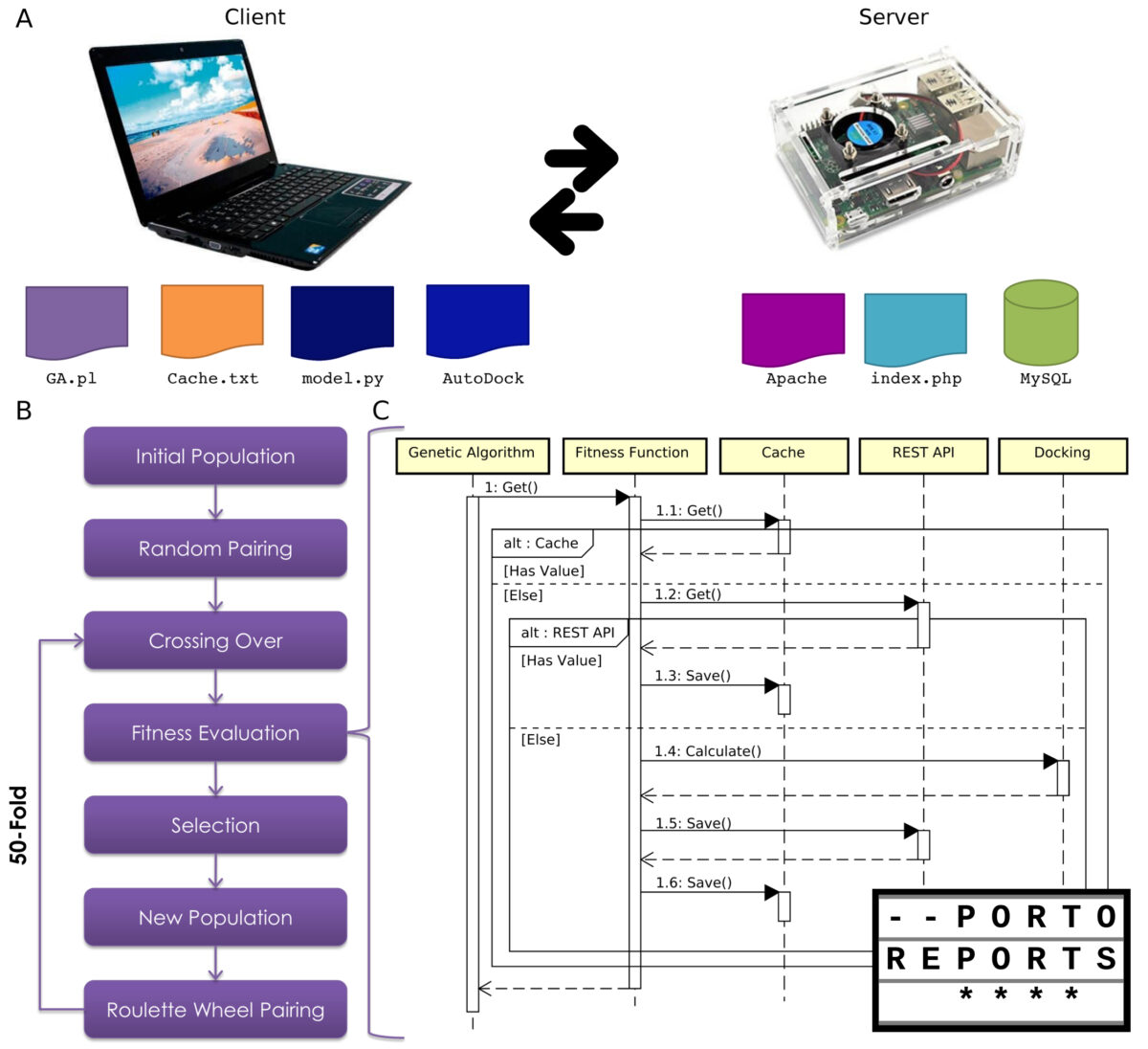
This system could be considered as a frugal innovation, due to the reuse of previous resources. I took advantage of the genetic algorithm developed for antimicrobial peptides and adapted it to use molecular docking as the fitness function. However, the main innovation was the use of a raspberry pi computer as a server. Interestingly, this feature arose from a failure on my notebook: it randomly stops to work and then a restart is required. Therefore, how I can recover all the data? Fortunately, I had the raspberry pi, which could act as a server, despite its computational power. Thus, with this client-server architecture, the system increase in performance and more than 70,000 peptides could be screened.
However, what is the main idea behind this project? Firstly, it should make clear that this was a very preliminary study, based on computer simulations. As well other virtual screening studies, the main target was the viral protease, which is pivotal in viral cycle, however, peptides have some advantages over other putative inhibitors, due to their plasticity, which turns them very versatile molecules, where other building blocks could be added to add functionality to the molecule.
In this context, taking the viral protease as the target, the molecule should enter the infected cell to reach the target. Therefore, if a molecule could inhibit the protease, but fail in entering the cell, the molecule probably will not work. In the case of peptides, this could be easily fixed by adding a cell penetrating peptide at one of the terminals.
Therefore, the two identified peptides (HHYWH and HYWWT) should a piece on this puzzle, but there is more to be discovered. The main question is if they really bind to the protease and upon binding if they inhibit or are just cleaved by the protease. Depending on what happens, from my point of view, different strategies for engineering a peptide drug could be used: firstly, in case of inhibition, the peptide should be linked to a cell penetrating peptide; and secondly, in case of cleavage, a toxin could be designed to kill the infected cells, by a combination of a four-domain peptide, including a toxin, the peptide, a toxin inactivating sequence and a cell penetrating peptide.
This clearly shows how preliminary the data is. Besides, there are further steps prior to approval for human use, including in vitro and in vivo assays. But we hope that this study could help in solving this critical scenario. By now, with the development of vaccines, we are close to the end, however, the data from this article, as well as the virtual screening system, could be useful for future pandemics.
Quality assessment:
Originality ☆☆☆☆✭
Rigor ☆☆☆☆✭
Significance to the field ☆☆☆☆☆
Interest to general audience ☆☆☆☆☆
Quality of writing ☆☆☆☆✭
Overall quality of the study ☆☆☆☆✭
Reference
Porto (2021) Virtual screening of peptides with high affinity for SARS-CoV-2 main protease. Computers in Biology and Medicine, vol 133, 104363. https://doi.org/10.1016/j.compbiomed.2021.104363